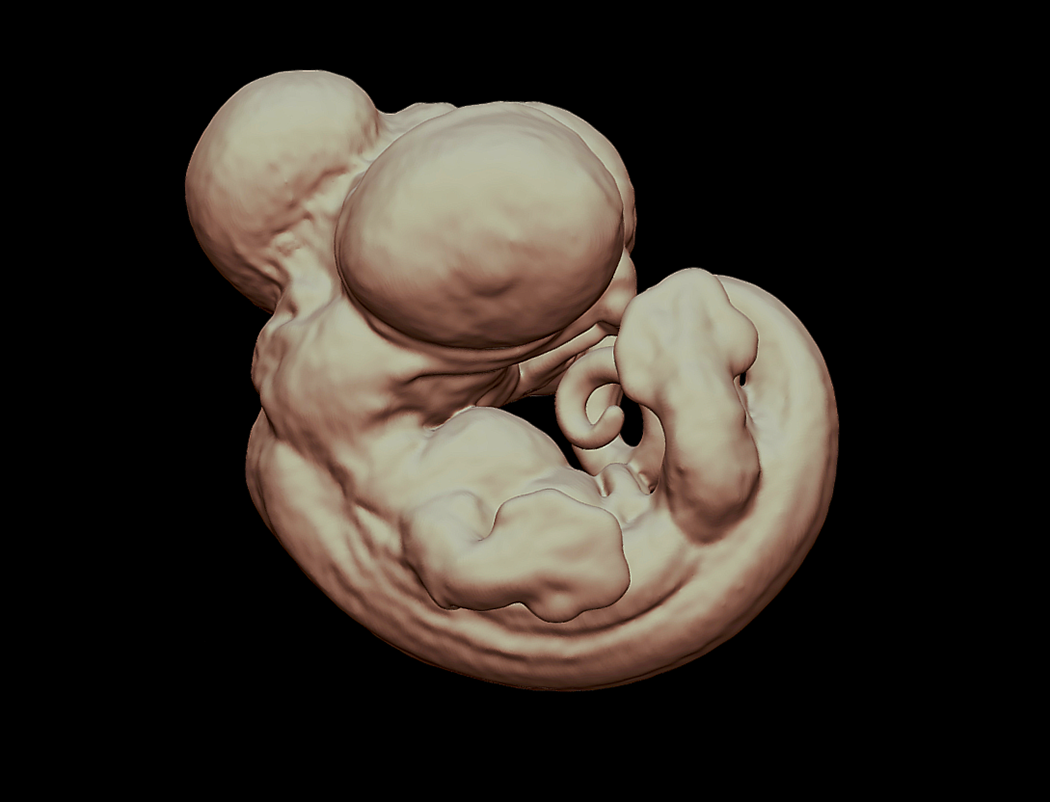
Three-dimensional Staging Series of Anolis sagrei
The following data are part of the presentation by Lilian Epperlein at the 2021 annual meeting of the Society of Integrative and Comparative Biology. If you have questions or would like more complete access to these data, please contact the authors.
With the rapid rise of Evo-devo and the concurrent development of new imaging techniques, comparative studies of development have accelerated over the last decade. Squamates, lizards and snakes, lack a traditional experimental model species for developmental investigations, yet are important for understanding fundamental evolutionary questions because of their remarkable diversity. However, several squamate species have growing communities of biologists building new resources for comparative and experimental studies of lizard development. Creation of detailed embryological atlases for these species will help promote their advancement. In the past, scientists have had to rely on destructive methods to take apart biological samples which allows them to visualize the three-dimensional (3D) anatomy. However, X-ray computed tomography, CT scanning, allows for sub-10 micron, non-destructive imaging of vertebrate embryos. This technique allows not only the ability to analyze the hard tissues, but also, with the help of chemical counterstains, the ability to differentiate among soft tissues. I created a detailed, 3D embryological atlas of the model lizard species, Anolis sagrei, using micro-CT scanning. For a subset of stages I reconstructed the development of both hard and soft tissues, such as bone, muscle, and neural tissues. All reconstructions were performed in the free software package 3D Slicer. This anatomical atlas will be essential for future research on Anolis development, will build a comprehensive understanding of the embryonic development of anoles, and may provide a reference for people investigating the development of other lizards.